Retinol as electron carrier in redox signaling, a new frontier in vitamin A research
Introduction
Carotenoids and retinoids are a large family of biomolecules with diverse functions that occur in all three domains of life (1). By far the largest within this group of compounds are the pigments, counting by the hundreds and familiar to us as delightful colors (2). Plants use these as visual signals to advertise fruit ripening, to attract pollinators and seed dispersers (3), among endless other tasks. Insects employ them for camouflage (4), birds display them in their plumage for mate selection (5), fishes decorate their flanks with them to facilitate swimming in formation (6). While these pigments are the most diverse in numbers, a more circumscribed group of carotenes associated with photosynthesis (7) supersede them by sheer biomass. Chlorophyll-based photosynthetic systems employ carotenes as light-harvesting antennas to broaden the range of usable photons (8,9), but also as shields against excessive illumination (10,11). And a single retinoid, retinaldehyde, is the chemical engine of bacteriorhodopsins (12) and animal rhodopsins (13) alike. The former drive proton pumps for ATP generation (14), ion pumps for reverse osmosis (15), or sensors for phototactic responses (16), whereas rhodopsins are the universal chromophores of vision (17,18).
The ability to monitor the physical world is of fundamental importance to all organisms. Carotenoids and retinoids are a vital part of this system. The key structural feature enabling carotenoids and retinoids to interact with diverse forms of electromagnetism is their linear system of conjugated double-bonds (19). π-electrons of this polyene absorb photons at wavelengths matching their intrinsic excitation spectra. Carotenoids (and retinoids) are constructed by concatenation of isoprene subunits. Hence plants can adapt carotenoids to a wide range of wavelengths by extending or shortening their polyenes through the addition or subtraction of isoprene units. A bathochromic effect is also obtainable by converting select single to double bonds by desaturases (4,20) and, since this step is reversible via saturases, modifying the polyene in this manner theoretically makes for real-time dynamic tuning systems. A retinol saturase was recently described by Moise et al. (21). Electro-philic or -phobic substitutions at the ends of the polyene alter the orbital energy levels of the electrons, creating further opportunities for fine-tuning of light absorption spectra. Physically embedding carotenoids within proteins, or ligating retinaldehyde with specific opsin carriers (22), is another common way of altering the absorption characteristics. Combinations of these strategies permit the carotenoid/retinoid pigment family to cover virtually the entire visual light spectrum, spanning from infrared to blue-green, and expanding even to the near-UV.
The common purpose of carotenoids and retinaldehyde is the conversion of energy from one form to another: light to chemical energy, light to nerve impulse, light to electrons, electrons to heat, and so on. Among the retinoid family, retinaldehyde alone was thought to possess the property of energy conversion, but vitamin A alcohol (retinol) was recently found capable of handling electrons as part of a specific redox-chemical reaction that controls mitochondrial energy homeostasis (23,24). In essence, two protein kinase C (PKC) isoforms, δ and ε, require retinol as indispensible co-factor for their function in mitochondria (25,26). In this organelle, activation of PKCs is not mediated by the classic diacyl-glycerol (DAG) second messenger (27), but is accomplished by an alternate redox mechanism in which retinol participates as an electron transfer agent (24,28-30). Since both PKC isoforms regulate the pyruvate dehydrogenase complex (PDHC), although opposing each other yin-yang style (25), retinol is part of a signal network that regulates the conversion of glucose to acetyl coenzyme A (CoA), and as such is fundamental to animated life. The PKC δ/ε-PDHC axis is crucial for the balanced flux of fuel entering the Krebs cycle in virtually all respiring mammalian cells, and thus retinol plays a critical role in containing the respiratory capacity within safe borders. It is becoming increasingly clear that harmful reactive oxygen species emanating from an overtaxed electron transfer chain (ETC) underlie a welter of metabolic disorders, impacting diabetes, cancer, cardiovascular disease, neurodegenerative disorders, and aging (31,32). The subject of this review is the emerging role of retinol as an electron carrier. In this vein, retinol performs a narrowly specialized task that at first glance may appear incompatible with the biological function of other retinoids, yet fits neatly into the electronic capabilities of the carotenoid/retinoid superfamily.
Physiological role of vitamin A, independent of retinaldehyde and retinoic acid (RA)
Vitamin A was long thought to lack intrinsic biological activity. Its sole function was believed to be precursor of various bioactive forms, notably retinaldehyde for vision (18) and RA for transcriptional regulation (33,34). However, vitamin A is uniquely defined as the alcohol form, retinol, and thus is distinct from other retinoids in its chemical structure and, owing to the discovery of the involvement in mitochondrial signaling, its biological function as well. A reluctance to accept bona fide biological activity for retinol can be attributed to the need for a new paradigm that would account for an intrinsic function that is radically different from the reigning retinoid acid paradigm. RA works primarily as a paracrine hormone (35). In accord, RA is elicited by one group of cells to signal the execution of a transcriptional program by another, receptive, population of cells. RA signaling is usually narrowly controlled in time and space, within a developmental field, or a regenerating tissue, for instance (36,37). To make this system work, RA receptors have evolved to high affinity in the sub-nanomolar range. Thus RA concentrations are usually exceedingly low, excess being promptly degraded by specific enzymes, exemplified by CYP26, that are strategically placed to prevent RA escaping from a designated developmental field (38). Moreover, RA production is transient.
The evolving retinol paradigm does not share these classic attributes of hormones. Retinol is distributed via the circulation to tissues and cells at all times. In healthy animals, the liver maintains retinol in plasma at an unwavering concentration of 1.5−2 micromoles (39). Moreover, the uptake of retinol occurs via redundant, though controlled, processes assuring that it is available to cells in relative abundance. This is the precise antithesis of a hormone. In distinction to RA, retinol does not activate the target molecules to which it binds, the members of the PCK and Raf families, for instance (29). Instead, binding of retinol primes these molecules to become responsive to redox-mediated activation signals that present an alternative to the classic, retinol-independent second-messenger pathway (40). In the case of PKCδ, which inhabits the mitochondrial intermembrane space, this signal is given by a locally activated oxidizing agent, cytochrome c3+ (24). However, without retinol this kinase activation process is impeded.
Brief history of vitamin A
Vitamin A was discovered during nutritional studies designed to define the minimal caloric requirements for raising chicks and rodents. However, in addition to the basic staples (carbohydrate, fat and protein) a number of low-abundant small organic compounds were found that, while not contributing any calories, were nevertheless essential for normal growth. One such accessory factor, fat-soluble A, was isolated from milk and later became known as vitamin A (41,42). Paul Karrer solved the chemical structure in 1932 (43), and a trio of two Dutchmen, Arens and Van Dorp (44), and a German, Isler (45) confirmed the structure by organic synthesis. Although early research results emphasized the growth-promoting qualities, an enormous complexity of vitamin A biology would soon follow. Among many such studies, none revealed the multiplex nature of vitamin A more eloquently than Wolbach and Howe’s systematic histopathology of rats reared on a vitamin A deficient diet (46). Their description of “Atrophy of many glands, arrest of growth, emaciation, and replacement of many different epithelia by stratified keratonizing epithelium actually characterize fat-soluble A avitaminosis” sums up the broad range of developmental defects, outside malformations of the eye, that had gone unnoticed to this point. Although it took another 50 years to come up with mechanistic explanations, such as those enshrined in the RA paradigm, the full complexity of vitamin A action is underappreciated to this day. For instance, while the causes of defective vision in vitamin A deficiency are now well understood, the equally dramatic loss of olfactory sensation observed by Wolbach and Howe (46) remains unexplained (47). When RA arrived on the scene and the 1920s nutritional studies were repeated, the power of RA to reverse many symptoms of the vitamin A deficiency was noted. However, several aspects of immune system dysfunction were not fully corrected with RA, but required vitamin A for reversal (48). It should be noted that vitamin A is converted in the body to RA, but the converse is not true.
Many arguments can be made that vitamin A is more than just a precursor, as it is often said, of bioactive molecules, such as retinaldehyde in vision, or RA in gene transcription. For instance, while it is true that retinoic acid receptors [RARs and retinoic acid X receptors (RXRs)] are ubiquitously expressed, and hence most cell types evidently use RA, the majority of these consumers do not synthesize RA. Nevertheless, most cells including the RA non-producers, harbor the biochemical machinery to take up vitamin A from the circulation via specific intracellular transporters, the cellular retinol binding proteins (CRBP) (49), to convert it for storage into retinyl esters by dedicated acyl transferases [lecithin retinol acyltransferase, LRAT (50) or acyl-CoA retinol acyltransferase, ARAT (51)] and to retrieve free retinol on demand by retinyl-esterases (52). Moreover, many cell types metabolize retinol into hydroxylated derivatives, including 14-hydroxy-retro-retinol (14-HRR) (53) and 13,14-dihydroxy-retinol (DHR) (54) which have in vitro growth-promoting qualities broadly similar to that of retinol, although it is unclear whether they act by a similar mechanism or what their biological relevance might be. Another retinol metabolite, anhydroretinol (AR) (55), was discovered owing to its capacity to compromise cell survival. Both 14-HRR and AR, along with retinyl esters, are evolutionarily-conserved in insect cells, suggesting physiological relevance of these compounds (and of the enzymes that generate these), although specific uses have not been investigated (56). Since RARs or RXRs emerged at the vertebrate/invertebrate boundary it is unlikely that 14-HRR or AR act as transcriptional co-activators. While these findings hint at the existence of an unknown vitamin A physiology they do not per se argue that unmodified vitamin A is bioactive. Arguments in favor of intrinsic bioactivity stem from the discovery of a large class of intracellular receptors of vitamin A, namely the serine/threonine PCK and Raf kinases (29).
Vitamin A (retinol) as cofactor of PKCs and cRaf
As a result of a fishing expedition with a retinoid-affinity matrix we deduced that the PKC and Raf kinases might be target molecules of vitamin A because several family members bound specifically to retinoid-coated beads. Focusing initially on cRaf, and later on PKC, we traced the retinol-binding site to the cysteine-rich domain (CRD) that these two proteins share in common. cRaf contains one such CRD whereas PKC contains two. In fact all other members of the Raf and PKC families possess retinoid-binding sites associated with at least one of their CRDs, although the novel PKCs (δ and ε) have sites on both CRDs. The binding affinities of retinol for CRDs range from 20 to 95 nm. These values are at least two orders of magnitude lower than the (sub-nanomolar) affinity of RA for RARs. On the other hand, the estimated PKC and Raf affinities for retinol are similar to those of extracellular and cytoplasmic RBPs (RBP and CRBP, respectively), suggesting that they are in equilibrium with these major retinol transporters. Therefore, in nutritionally healthy animals at least a proportion of the Raf and PKC isoforms will always be complexed with retinol. It should however be understood that the recorded affinities relate to recombinant CRD fragments, and that the true values for the native proteins in their cellular environments may be substantially different.
CRaf and PKC CRDs were found to bind other vitamin A metabolites with similar affinities as retinol, including RA, the hydroxylated derivatives 14HRR and 13,14-DHR, and AR, as well as a host of synthetic retinoids (28). Because of this lack of selectivity doubts arose that binding was non-specific and hence biologically irrelevant. However, an analysis of the retinol contact sites on the PKC αC1A CRD by systematic point mutagenesis revealed a defined binding pocket that was preserved to high homology in other retinol-binding CRDs, but was dissimilar in non-binding CRDs. Moreover, the naturally non-binding αC1B domain converted to a retinol-binding domain when its three incongruous contact amino acids (Thr-7, Tyr-8, Tyr-22) were replaced by the αC1A consensus residues, Phe-7, Phe-8, or Trp-22 (57). In silico docking studies confirmed the general layout of the retinol-binding pocket, with a head-first orientation of retinol, but crystallographic confirmation is still outstanding. Indeed, the interaction of the two hydrophobic contact amino acids mentioned above with the β-ionone ring of retinol would neatly explain why other vitamin A metabolites have similar binding capacity as retinol itself: they possess the same β-ionone structure, while being chemically distinguished by modifications of their polyene tails. The latter is assumed to protrude from the pocket, contributing little to binding affinity but affecting their biological function. A dramatic example is AR that induces cell necrosis, instead of supporting viability (like retinol does) (58).
Localization and function of PKCδ and PKCε isoforms in mitochondria
Both PKCδ and PKCε translocate from the cytosol to mitochondria, but whereas PKCδ migrates to the intermembrane space, PKCε is transported into the matrix. Despite their different locations, or perhaps by design, the two isoforms signal to a common target, the PDHC (25) (Figure 1). However, the nature of their signals is different: PKCδ transmits a stimulatory signal, and PKCε an inhibitory one. These conclusions were reached from the analyses of the phosphorylation patterns of interacting proteins in intact cells. Thus, the phosphorylation of Thr505 on PKCδ, which signifies an active kinase, correlated with de-phosphorylation of the Ser293-site of the E1 regulatory subunit of PDHC, which indicates increased enzyme activity (23). Conversely, activation of PKCε increased E1 subunit phosphorylation, hence decreased PDHC activity (25). Our results challenge published reports that proposed the reverse polarity (59), while agreeing with PKCδ and PKCε as opposing each other. These authors assumed inactivation of PDHC by PKCδ and activation by PKCε. However, their conclusions were based on cell-free assay systems without stringent validation in vivo, and hence draw considerable skepticism.
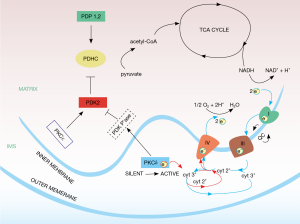
Although the correlations between PKCδ and PDH E1 phosphorylation patterns (60,61) identified this kinase as positive regulator of the PDHC the actual signal path was indirect. First, PKCδ is a phosphotransferase, but PDH became dephosphorylated when the PKCδ kinase was active. This implied the participation of an intermediary phosphatase. In fact, two pyruvate dehydrogenase phosphatases, PDP1 and 2, are known to directly control the PDHC E1 subunit (62,63), but both could be excluded from the PKCδ signal chain proper. A required phosphatase that is activated by PKCδ has not yet been identified. Second, the pyruvate dehydrogenase kinase isoform 2 (PDK2) (64), was identified as the intermediate target of the PKCδ signal path (23). Along with three other PDK isoforms, in its active, phosphorylated form PDK2 is a known suppressor of PDHC, acting by phosphorylating the PDHC E1 subunit (61). On the other hand, PDK2 is opposed by pyruvate dehydrogenase phosphatase 1,2 (PDP1,2) which dephosphorylates PDHC E1. Thus, PKCδ seems to function by activating PDK2 via the hypothetical, PKCδ-dependent PDK2-phosphatase. When PDK2 is neutralized by dephosphorylation, PDP1,2 is unopposed to activate the PDHC.
Wherever a phosphatase, there is usually a compensating kinase, and vice versa. The kinase that activates PDK2 has been elusive, but our unpublished results indicate that activation of PKCε increases the phosphorylation status of PDK2, as defined by 2-D gel electrophoresis (Gong and Hammerling, unpublished). Hence these data point to PKCε as the PDK2 kinase. While the connection to PDK2 needs to be confirmed, the evidence is strong that activation of PKCε suppresses PDHC activity (25). The participation of PDK2 in this inhibitory branch of the PKC signal network would fit well into the overall framework (Figure 1).
Zinc-fingers as key elements controlling PKC activation
In his pioneering work, Dr. Nishizuka (27) established the activation of PKC by diacyl-glygerol second messenger, and later added an alternate mechanism involving redox stress (65). Despite descriptive evidence for other family members, notably cRaf, this redox mechanism raised doubts in the minds of biologists as to the physiological purpose, as well as of chemists who worried about vulnerability of this cysteine-rich molecule towards randomly oxidizing agents. On the other hand, the activation domains of PKCs and Raf, which are synonymous with the aforementioned CRDs, are organized into zinc fingers (66-68). These folds are generally believed to stabilize tertiary protein structure as long as reducing conditions prevail, as they normally do in intact cells, but CRDs dissociate when oxidizing conditions arise systemically or locally, allowing the respective proteins to change function by rearranging form. Bacteria adopted this paradigm for flexible control of protein activity, as described for the prototypic Hsp33 chaperone (69,70).
Zinc-fingers are devices to maintain PKCs in the closed, that is: enzymatically inactive, form. But it is attractive to view these structures as hinges that open in order to effect kinase activation. In fact, the transition from inactive to active enzyme classically involves large-scale protein unfolding to expose targeting structures and various binding sites for substrate, ATP, and, for some isoforms, Ca2+ (71). Activation is initiated by the interaction of the CRD with second messenger, the long-chain fatty acids of DAG bestowing a hydrophobic patch to the PKC. This modification is believed to allow the protein to translocate from cytoplasm to preordained sites in membranes, where step-wise unfolding of the kinase occurs under the influence of the hydrophobic milieu. However, we challenge this sequence of events and propose that second-messenger binding triggers the unfolding of the CRD, and that newly exposed recognition sites on the PKC subsequently mediate the translocation to membranes (26).
While analyzing Nishizuka’s redox stress mechanism we discovered that vitamin A needed to occupy the CRD in order to effect kinase activation by redox stress initiated by hydrogen peroxide (28). The PKC zinc-fingers are constructed by two sets of three cysteines and one histidine, each chelating one zinc ion (67). This dual zinc-finger fold is highly susceptible to oxidation, owing to the proximity of several cysteine residues. Moreover when vitamin A binds in the immediate vicinity of the zinc-coordination center it might tag particular cysteines for preferential oxidation. On theoretical grounds the loss of negative charges of the cysteine-sulfhydryl anions (which is tantamount to oxidation) must lead to the collapse of zinc-coordinated structures. We hypothesized that the ensuing local structural change triggers the large-scale protein remodeling referred to above (71). We further postulated that DAG binding might also cause disassembly of the zinc-finger (26). While there is no precedent for such a mechanism, it is conceivable that DAG disrupts the symmetry of the zinc-coordinated fold. The stability of the latter depends not only on the presence of cysteine-sulfhydryl anions and histidinyl imine anions, but on their precise placement at the corners of a perfect tetrahedron. If DAG were to dislodge even one of the four ligands the spherical ionic field might irreversibly collapse leading, like oxidation, to an open CRD structure.
Using native and recombinant proteins we obtained supporting evidence as follows: (I) the active form of PKC, isolated from stimulated cells, contained half as much Zn2+ than the dormant form (72); (II) following mild oxidation of the CRD, or stimulation with phorbol myristate acetate (PMA) (a pharmacomimetic of DAG), the binding affinity of Zn2+ changed from high to low, a clear indication of uncoordination of the zinc-finger; (III) the loss of Zn2+ coordination was accompanied by the change in the NMR HSQC spectrum from a well-dispersed pattern of cysteine residues to a random pattern, indicating loss of tertiary structure; (IV) the substitution of the bacterial HSP33 zinc-finger domain for the mammalian PKCεC1B zinc-finger conferred sensitivity to phorbol ester stimulation while preserving HSP33 chaperone activation by oxidation (26). In summary, PKC zinc-fingers, long considered as static stabilizers of tertiary structure, are in fact dynamic hinges, like their bacterial orthologs (69), whose conformation can be controlled by redox chemistry or lipid agonist binding.
Activation of PKCδ in mitochondria: a new paradigm
PKCδ resides in the intermembrane space as a tetrameric complex, comprised of the following partners: the signal adapter protein, p66Shc (73), that interacts via its SH2 domain with phosphotyrosine Y332 of PKCδ (74); cytochrome c, that binds to p66Shc via hydrophobic interaction with glutamate residues E332 and E333 of p66Shc (75); vitamin A, that occupies the CRD binding pocket within the PKCδ C1B zinc-finger, as described above (Figure 2). Integrity of the tetrameric configuration is vital for PKCδ activation in mitochondria, since genetic ablation of PKCδ or p66Shc, or removal of vitamin A cofactor prevented signaling. Likewise, mutations of any of the three interaction sites described above led to inaction (24). However, in each case the blockade was overridden by phorbol ester stimulation, indicating that as long as physically present although not properly integrated into the signalosome, PKCδ was enzymatically intact and was capable of forward signaling to PDHC. The importance of the retinol cofactor for signaling was underscored by the use of an engineered PKCδ that lost the retinol-binding site and as a result was inactive in mitochondria (24). As with the other genetic inactivations, this loss mutation was overridden by phorbol ester.
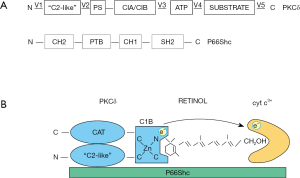
In mitochondria the activating signal was given by oxidized cytochrome c3+, implying a redox mechanism (a concrete form of Nishizuka’s “stress” signal). This mechanism is without precedent, and is distinguished from the classic DAG second messenger operating outside mitochondria, yet initiates a similar unfolding cascade as DAG, as alluded to above. Note that cytochrome c is restricted to healthy mitochondria and has, owing to its fixation to p66Shc, the potential to interact with PKCδ in a site-directed manner (see Figure 2).
The presumptive target of cytochrome c3+ is the PKCδ C1B zinc finger, and the presumptive activating reaction is the oxidation of zinc-finger cysteine(s) yet to be defined, leading to loss of zinc-coordination and initiating large-scale remodeling of the PKCδ protein. Oxidation was inferred from the observed conversion of high-affinity coordinated Zn2+ ions to low-affinity bound forms. The primary product of this chemical reaction is not known, but is likely to involve a cysteinyl sulfine intermediate which, by reaction with a neighboring cysteine, may resolve into stable cysteine-disulfide (Zn2+ might be retained in the CRD in low-affinity bound form as sulfide). Oxidation involves the transfer of a pair of electrons from cysteine sulhydryl anions to cytochrome c. Using isolated mitochondria, we verified that oxidation was carried out by cytochrome3+, but not by cytochrome C2+ (24).
Retinol as electron transfer agent
The heme of cytochrome c3+ absorbs one electron at a time. Therefore the left-behind electron of the pair to be transferred to cytochrome c3+ inevitably forms an intermediary radical, which needs to be stabilized until cytochrome c3+ is regenerated from cytochrome c2+ by cytochrome c oxidase. However, the transfer of electrons between proteins that are not in VanDerWaal’s contact (like the passage from PKCδ to cytochrome c3+) is exceedingly slow, and hence requires an electron bridge. PKC-bound retinol is proposed to perform this function. Close similarities exist with electron transfer in the ETC. This process is dictated by one-electron chemistry that mediates directional passage of electrons from NADH-CoQ reductase (complex I) or succinate-CoQ reductase (complex II) to CoQH2-cyochrome c reductase (complex III). Coenzyme Q greatly accelerates this step (76). Note that coenzyme Q toggles between different chemical forms, ubiquinone ↔ semiquinone radical ↔ ubiquinol. We propose that PKCδ employs retinol for the analogous purpose of enabling safe and efficient electron transfer to cytochrome c3+ (Figure 2). However in distinction to coenzyme Q retinol may not need to change its redox state, as the transitory electron may be stabilized by the π-electron system. While the details have to be worked out, it is known that an intact electron system is required. As the experiment with 11,12-dihydro-retinol showed, the reduction of even one double to a single C-C bond abolished PKCδ activation in the mitochondrion (24).
Relevance of PKCδ and vitamin A for metabolic disease
The PDHC catalyzes the conversion of pyruvate to acetyl-CoA, the last step of glycolysis. As our evidence shows, the PKCδ signaling module positively regulates the PDHC. A survey of the literature reveals that PKCδ-signaling pathways are directly involved at several points in energy metabolism. In particular, PKCδ affects glucose transport and utilization (77), gluconeogenesis and insulin secretion (78,79), insulin signaling (80-82), insulin resistance (83), and cellular oxidative stress (84). The latter is a key factor believed to drive the metabolic syndrome (31,85,86). In addition, lipoprotein metabolism and hepatic liponeogenesis were linked to PKCδ (87). With such broad relevance to regulation of energy homeostasis it comes as no surprise that genetic or pharmacologic manipulations impairing any of the four protein components of the PKCδ signalosome undercut the mitochondrial balance, profoundly affecting life span (88,89), obesity, and especially type 2 diabetes (90). Specifically, differences in PKCδ protein expression levels strongly influenced glucose utilization in mice. In excess, PKCδ predisposed mice towards obesity and metabolic syndrome (91). It seems that chronic activation of the PKCδ signaling pathway leading to preferred glucose fuel use at the expense of fat (which goes into storage) could be a factor in increased adiposity. The flip side to proneness of the PKCδ over-expressing mice towards metabolic syndrome was that PKCδ knockout mice were resistant to diet-induced obesity and obesity-related disorders (91) owing to lesser reliance on glucose over lipid metabolism, accompanied by diminished oxidative stress. PKCδ-null mice were lean, displayed low abdominal fat mass and likely had slower metabolic rate (91). Genetic ablation of p66Shc in mice largely phenocopied PKCδ-null mice (92,93), likely because p66Shc is required for PKCδ-mediated control of the PDHC. Furthermore, since both PKCδ and p66Shc were linked to the generation of ROS in mitochondria (75,84) the resistance of the PKCδ and p66Shc knockout mice towards metabolic disease could be ascribed to the reduced load of harmful oxygen radicals.
Retinol is a component of the PKCδ signalosome, the presence of which is mandatory for PKCδ activation, as described above. Omitting vitamin A from cell cultures compromised the PKCδ/PDHC signal path (23), but limiting dietary vitamin A in mice is impractical. However, lowering vitamin A in the circulation was accomplished by reducing the vitamin transporter, retinol binding protein (RBP), with drugs (94-96), or by genetic inactivation of the RBP4 gene (97). Both interventions resulted in improved insulin-resistance and glucose tolerance in diet-induced obesity (97). The fact that RBP knockout mice have limited access to hepatic vitamin A stores (98) suggested that the RBP knockout phenotype was linked to an impairment of the PKCδ signalosome, since the lack of adequate vitamin A was likely to reduce the proportion of retinol-bound PKCδ available for redox activation in mitochondria and hence reduce reliance on glucose for oxidative phosphorylation (OXPHOS). The converse was also true: mice expressing a human RBP4 gene were obese and displayed increased susceptibility to insulin resistance. Similar results were obtained when mice were repeatedly injected with holoRBP protein (97). In either case, the extra amount of retinol available to tissues could skew PKCδ signaling, leading to increased PDHC activity. Like the above-mentioned chronic excess of PKCδ that promotes obesity and increases the risk of diabetes, the overabundance of PKCδ cofactor could shift energy generation towards glucose fuel, away from fat utilization.
However, alternate explanations of how holoRBP affects glucose metabolism have been proposed. Thus, the stimulated by retinoic acid 6 (STRA6) receptor of holoRBP facilitates vitamin A uptake (99). In addition, when stimulated by holoRBP, STRA6 engages the JAK2/STAT5 cascade which controls several parameters of insulin signaling. Excessive STRA6 signaling which occurs in obesity due to pathologically elevated holoRBP levels was linked to insulin resistance (100,101). To the contrary, STRA6 knockout compared to WT mice displayed increased insulin sensitivity (102) attributable, at least in part, to the attenuation of JAK2/STAT5 signaling. While neither the PKCδ nor the STRA6/JAK2/STAT5 pathways alone can account for glucose utilization, together they might function as two cooperative branches of a broader signal system that balances glycolytic energy expenditures.
A third explanation for the increased risk of obesity and metabolic syndrome by overabundance of holoRBP was advanced by Moraes-Vieira et al. (103). These authors invoked a cytokine-like action of RBP that provoked an inflammatory response in adipose tissues via activation of antigen presenting cells and infiltration of CD4 T cells. Conceivably, all three factors: PDHC dysregulation, STRA6/Jak2/Stat5 signaling and inflammation via of Toll receptor 4 activation (104) conspire in situations of pathologically high holoRBP levels to promote metabolic disease.
How can chronic vitamin A excess lead to pathology? The action of two inhibitory retinoids, AR (55) and fenretinide (105) may be informative. In cell culture experiments, these retinoids caused mitochondrial stress and necrotic cell death (25,58,106-109). Both retinoids are known to bind the PKCδ activation domain with similar affinity as retinol (28), but inexplicably they caused the hyper-activation of mitochondrial PKCδ (25) leading to cell death. This was reflected in one study by high levels of ROS (58), while in another AR-treated cells became depleted of ATP to a degree that they could no longer survive (107). The paradox, why AR (or feneretinide) can substitute for retinol as activating cofactors of PKCδ, but why this mode of activation leads to cell necrosis, remains unresolved. It is noteworthy that supranormal retinol levels (above 2 micromoles) are also toxic, as the inverted U-shaped retinol dose responses indicate (25). The common denominator may be uncontrolled ROS production stimulated by the inordinately high PDHC activity. The important realization is that both the concentration and the type of retinoid determine the function of PKCδ.
The question then arises how PKCδ cooperates in physiological settings with retinol and, if occupancy of the retinol-binding site is the defining factor, how PKCδ is loaded with retinol. Retinol is membrane-permeable and partitions in vitro from the medium into cells mainly by diffusion (110). Occupancy of PKCδ is therefore determined by the extracellular retinol concentration. The in vivo situation is more complicated, since retinol is handled by two chaperones, RBP and CRBP. Retinol dissociates from the holoRBP/transthyretin (TTR) complex and spontaneously partitions across the plasma membrane where it is picked up by CRBP. Alternately, or in parallel, the STRA6 receptor facilitates the dissociation of retinol from holoRBP and transports it into the cell (111). Whether dictated by spontaneous diffusion (110) or facilitated by STRA6 (111), the rate of retinol uptake and degree of PKCδ saturation are determined by the concentration gradients and the relative affinities for retinol of the RBP/CRBP/PKCδ chain. Respective retinol-binding affinities were measured so far only in vitro with full-length recombinant proteins, or protein fragments. Accordingly, RBP and CRBP1 binding constants are ca 15 nM (112), whereas that of the isolated PKCδ C1b domain [relevant for mitochondrial function (24,25)] is 65 nM (28). To the extent that these values reflect the in vivo reality, the binding affinity of PKCδ is at, or even below, those of the RBP and CRBP1 transporters. Since the PKCδ retinol occupancy rate is dictated by mass action an upward shift in the extracellular vitamin A supply, as is the case with hRBP transgenic expression for instance, would translate into a higher PKCδ occupancy rate and, therefore, a larger activatable pool of PKCδ. Likewise, increasing the PKCδ expression level, as happens in C57Bl/6 compared to 129 mice (91), could increase the pool of retinol-primed PKCδ, given that intracellular retinol levels will not be limiting. Of course the larger the supply of the PKCδ/retinol complex would be, the higher the activation rates of the PDHC and the ever-present risk of over-stressing the ETC.
Unresolved problems
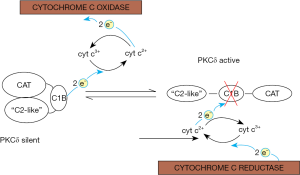
Our findings represent the beginnings of an intra-mitochondrial control system of aerobic glycolysis. To the extent that mitochondria of a given cell are separate entities the question arises how this signaling system is coordinated among these organelles. Other questions left unaddressed by this review are the interplay with the opposing PKCε signal pathway that is also dependent on retinol, and indeed the crosstalk with the myriads of extra-mitochondrial signals that control OXPHOS. Unresolved is also the problem of how PKCδ, once activated, is turned off again, as it surely needs to be, because continuous signaling is lethal (84). Considering the fact that PKCδ employs a redox-sensitive zinc-finger-fold for its activation and that redox-reactions are readily reversible, an attractive possibility is that PKCδ is inactivated by reduction. This might be accomplished via reversal of the polarity of the PKCδ-retinol-cytochrome c redox chain. Indeed, whenever cytochrome c2+ prevails, as would be the case in the resting state, the redox potential of this reductant could be high enough to restore the zinc-finger thereby returning the kinase to its inactive form (provided that a suitable chaperone would assist in refolding the protein). A mechanism wherein PKCδ oscillates between active (oxidized) and silent (reduced) forms would allow for real-time regulation of the PDHC (Figure 3). Retinol would be required to catalyze both the forward and reverse reactions. Still unclear is the nature of retinoid toxicity that arises when other retinoids, such as AR, substitute for retinol. However, if this finely tuned redox system were to be upset by a retinoid with different electronic properties (i.e., with different activation energies of their π-electrons), conceivably the forward, but not the reverse, reaction might be sustained, leaving the PKCδ stuck in the activated state which causes irreversible damage.
Outlook
True to predictions that their system of conjugated double bonds, like those of carotenoids, enables retinoids to interact with electromagnetism, retinol was shown to act as electron carrier in mitochondria. Retinol electronically couples the redox center of PKCδ (i.e., its zinc-finger-like activation domain) to cytochrome c, facilitating the activation of this kinase by a one-electron chemical mechanism. Since numerous isoforms of the threonine/serine family of kinases have homologous retinol binding sites in their activation domains it bears investigation whether the newly discovered biochemistry of retinol extends beyond mitochondria to other redox systems.
Acknowledgements
The author wishes to acknowledge generous grant support by Sloan-Kettering Institute for Cancer Research, New York, USA.
Footnote
Conflicts of Interest: The author has no conflicts of interest to declare.
References
- Goodwin TW. Metabolism, nutrition, and function of carotenoids. Annu Rev Nutr 1986;6:273-97. [PubMed]
- Grotewold E. The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 2006;57:761-80. [PubMed]
- Kevan PG, Baker HG. Insects as flower visitors and pollinators. Annu Rev Entomol 1983;28:407-53.
- Moran NA, Jarvik T. Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science 2010;328:624-7. [PubMed]
- Brush AH. Metabolism of carotenoid pigments in birds. FASEB J 1990;4:2969-77. [PubMed]
- Maoka T. Carotenoids in marine animals. Mar Drugs 2011;9:278-93. [PubMed]
- Frank HA, Cogdell RJ. Carotenoids in photosynthesis. Photochem Photobiol 1996;63:257-64. [PubMed]
- Ritz T, Damjanović A, Schulten K, et al. Efficient light harvesting through carotenoids. Photosynth Res 2000;66:125-44. [PubMed]
- Qin X, Suga M, Kuang T, et al. Structural basis for energy transfer pathways ub the plant PSI-LHCI supercomplex. Science 2015;348:989-95. [PubMed]
- Niyogi KK. Photoprotection revisited: Genetic and Molecular Approaches. Annu Rev Plant Physiol Plant Mol Biol 1999;50:333-59. [PubMed]
- Standfuss J, Terwischa Van Scheltinga A, Lamborghini M, et al. Mechanism of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5 A resolution. EMBO J 2005;24:919-28. [PubMed]
- Deisenhofer J, Epp O, Miki K, et al. X-ray structure analysis of a membrane protein complex. Electron density map at 3 A resolution and a model of the chromophores of the photosynthetic reaction center from Rhodopseu-domonas viridis. J Mol Biol 1984;180:385-98. [PubMed]
- Bridges CD. The rhodopsin-porphyropsin visual system. In: Dartnall HJ, editor. Photochemistry of vision. Handbook of sensory physiology. Heidelberg: Springer-Verlag, 1972:417-80.
- Lozier RH, Bogomolni RA, Stoeckenius W. Bacteriorho-dopsin: a light-driven proton pump in Halobacterium Halobium. Biophys J 1975;15:955-62. [PubMed]
- Stoeckenius W, Bogomolni RA. Bacteriorhodopsin and related pigments of halobacteria. Annu Rev Biochem 1982;51:587-616. [PubMed]
- Foster KW, Saranak J, Patel N, et al. A rhodopsin is the function-al photoreceptor for phototaxis in the unicellular eukaryote Chlamydomonas. Nature 1984;311:756-9. [PubMed]
- Wald G. Vitamin A in eye tissues. Nature 1933;132:316-7.
- Wald G. The molecular basis of visual excitation. Nature 1968;219:800-7. [PubMed]
- Hammerling U. The centennial of vitamin A: a century of research in retinoids and carotenoids. FASEB J 2013;27:3887-90. [PubMed]
- Linden H, Misawa N, Saito T, et al. A novel carotenoid biosyn-thesis gene coding for zeta-carotene desaturase: functional expression, sequence and phylogenetic origin. Plant Mol Biol 1994;24:369-79. [PubMed]
- Moise AR, Dominguez M, Alvarez S, et al. Stereospecificity of retinol saturase: absolute configuration, synthesis, and biological evaluation of dihydroretinoids. J Am Chem Soc 2008;130:1154-5. [PubMed]
- Sakmar TP, Franke RR, Khorana HG. The role of the retinylidene Schiff base counterion in rhodopsin in determining wavelength ab-sorbance and Schiff base pKa. Proc Natl Acad Sci U S A 1991;88:3079-83. [PubMed]
- Acin-Perez R, Hoyos B, Zhao F, et al. Control of oxidative phosphorylation by vitamin A illuminates a fundamental role in mitochondrial energy homoeostasis. FASEB J 2010;24:627-36. [PubMed]
- Acin-Perez R, Hoyos B, Gong D, et al. Regulation of intermediary metabolism by the PKCdelta signalosome in mitochondria. FASEB J 2010;24:5033-42. [PubMed]
- Gong J, Hoyos B, Acin-Perez R, et al. Two protein kinase C isoforms, δ and ε, regulate energy homeostasis in mitochondria by transmitting opposing signals to the pyruvate dehydrogenase complex. FASEB J 2012;26:3537-49. [PubMed]
- Zhao F, Ilbert M, Varadan R, et al. Are zinc-finger domains of protein kinase C dynamic structures that unfold by lipid or redox activation? Antioxid Redox Signal 2011;14:757-66. [PubMed]
- Nishizuka Y. Intracellular signaling by hydrolysis of phospholipids and activation of protein kinase C. Science 1992;258:607-14. [PubMed]
- Imam A, Hoyos B, Swenson C, et al. Retinoids as ligands and coactivators of protein kinase C alpha. FASEB J 2001;15:28-30. [PubMed]
- Hoyos B, Imam A, Chua G, et al. The Cysteine-rich Regions of the Regulatory Domains of Raf and Protein Kinase C as Retinoid Receptors. J Exp Med 2000;192:835-45. [PubMed]
- Hoyos B, Imam A, Korichneva I, et al. Activation of c-Raf Ki-nase by Ultraviolet Light. J Biol Chem 2002;277:23949-57. [PubMed]
- Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature 2001;414:813-20. [PubMed]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature 2000;408:239-47. [PubMed]
- Petkovich M, Brand NJ, Krust A, et al. A human retinoic acid receptor which belongs to the family of nuclear receptors. Nature 1987;330:444-50. [PubMed]
- Giguere V, Ong ES, Segui P, et al. Identification of a receptor for the morphogen retinoic acid. Nature 1987;330:624-9. [PubMed]
- Hofmann C, Eichele G. Retinoids in development. In: Sporn MB, Roberts AB, Goodman DS, editors. The Retinoids: Biology, Chemistry and Medicine. 2nd ed. New York: Raven Press, 1994:387-441.
- Tickle C, Alberts B, Wolpert L, et al. Local application of retinoic acid to the limb bond mimics the action of the polarizing region. Nature 1982;296:564-6. [PubMed]
- Thaller C, Eichele G. Identification and spatial distribution of retinoids in the developing chick limb bud. Nature 1987;327:625-8. [PubMed]
- Sakai Y, Meno C, Fujii H, et al. The retinoic acid-inactivating enzyme CYP26 is essential for establishing an uneven distribution of retinoic acid along the anterio-posterior axis within the mouse embryo. Genes Dev 2001;15:213-25. [PubMed]
- Blaner WS, Olson JA. Retinol and retinoic acid metabolism. In: Sporn MB, Roberts AB, Goodman DS, editors. The Retinoids: Biology, Chemistry, and Medicine, ed 2. New York: Raven Press,1994:229.
- Hoyos B, Acin-Perez R, Fischman DA, et al. Hiding in plain sight: uncovering a new function of vitamin A in redox signaling. Biochim Biophys Acta 2012;1821:241-7.
- McCollum EV, Davis M. The necessity of certain lipins in the diet during growth. J Biol Chem 1913;15:167-75.
- Osborne TB, Mendel LB. The relationship of growth to the chemical constituents of the diet. J Biol Chem 1913;15:311-26.
- Semba RD. On the 'discovery' of vitamin A. Ann Nutr Metab 2012;61:192-8. [PubMed]
- Arens JF, Van Dorp DA. Synthesis of some compounds possessing vitamin A activity. Nature 1946;157:190. [PubMed]
- Isler O, Huber W, Ronco A, et al. Synthese des vitamin A. Helvetica Chimica Acta 1947;30:1911-27. [PubMed]
- Wolbach SB, Howe PR. Tissue changes following deprivation of fat-soluble A vitamin. J Exp Med 1925;42:753-77. [PubMed]
- Garrett-Laster M, Russell RM, Jacques PF. Impairment of taste and olfaction in patients with cirrhosis: the role of vitamin A. Hum Nutr Clin Nutr 1984;38:203-14. [PubMed]
- Ross A, Hammerling U. Retinoids in the immune system. In: Sporn MB, Roberts AB, Goodman DS, editors. The retinoids: biology, chemistry and medicine. New York, NY: Raven Press, 1994:521-44.
- Ong DE, Chytil F. Specificity of cellular retinol-binding pro-tein for compounds with vitamin A activity. Nature 1975;255:74-5. [PubMed]
- Ruiz A, Winston A, Lim YH, et al. Molecular and biochemical characterization of lecithin retinol acyltransferase. J Biol Chem 1999;274:3834-41. [PubMed]
- Helgerud P, Petersen LB, Norum KR. Acyl CoA:retinol acyltransferase in rat small intestine: its activity and some properties of the enzymic reaction. J Lipid Res 1982;23:609-18. [PubMed]
- Chen CC, Heller J. Retinyl esterase activity of purified rat liver retinyl ester lipoprotein complex. Archives of biochemistry and biophysics 1979;198:572-9. [PubMed]
- Buck J, Derguini F, Levi E, et al. Intracellular signaling by 14-hydroxy-4,14-retro-retinol. Science 1991;254:1654-6. [PubMed]
- Derguini F, Nakanishi K, Hammerling U, et al. 13,14-Dihydroxy-retinol, a new bioactive retinol metabolite. J Biol Chem 1995;270:18875-80. [PubMed]
- Buck J, Grun F, Kimura S, et al. Anhydroretinol: A naturally occurring inhibitor of lymphocyte physiology. J Exp Med 1993;178:675-80. [PubMed]
- Grün F, Noy N, Hammerling U, et al. Purification, cloning, and bacterial expression of retinol dehydratase from Spodoptera frugiperda. J Biol Chem 1996;271:16135-8. [PubMed]
- Hoyos B, Jiang S, Hammerling U. Location and functional significance of retinol binding sites on the serine/threonine kinase, cRaf. J Biol Chem 2005;280:6872-8. [PubMed]
- Chen Y, Buck J, Derguini F. Anhydroretinol induces oxida-tive stress and cell death. Cancer Res 1999;59:3985-90. [PubMed]
- Churchill EN, Murreil CL, Chen CH, et al. Reperfusion-induced translocation of delta-PKC to cardiac mitochondria prevents pyruvate dehydrogenase reactivation. Circ Res 2005;97:78-85. [PubMed]
- Patel MS, Roche TE. Molecular biology and biochemistry of pyruvate dehydrogenase complexes. FASEB J 1990;4:3224-33. [PubMed]
- Patel MS, Korotchkina LG. Regulation of the pyruvate de-hydrogenase complex. Biochem Soc Trans 2006;34:217-22. [PubMed]
- Bowker-Kinley MM, Davis WI, Wu P, et al. Evidence for existence of tissue-specific regulation of the mammalian pyruvate dehydrogenase complex. Biochem J 1998;329:191-6. [PubMed]
- Holness MJ, Sugden MC. Regulation of pyruvate dehy-drogenase complex activity by reversible phosphorylation. Biochem Soc Trans 2003;31:1143-51. [PubMed]
- Popov KM, Kedishvili NY, Zhao Y, et al. Primary structure of pyruvate dehydrogenase kinase establishes a new family of eukaryotic protein kinases. J Biol Chem 1993;268:26602-6. [PubMed]
- Kikkawa U, Matsuzaki H, Yamamoto T. Protein kinase C delta PKC delta: activation mechanisms and functions. J Biochem 2002;132:831-9. [PubMed]
- Hurley JH, Newton AC, Parker PJ, et al. Taxonomy and function of C1 protein kinase C homology domains. Protein Sci 1997;6:477-80. [PubMed]
- Zhang G, Kazanietz MG, Blumberg PM, et al. Crystal Structure of the Cys2 Activator-Binding Domain of Protein Kinase C8 in Complex with Phorbol Ester. Cell 1995;81:917-24. [PubMed]
- Mott HR, Carpenter JW, Zhong S, et al. The solution structure of the Raf-1 cysteine-rich domain: a novel ras and phospholipid binding site. Proc Natl Acad Sci U S A 1996;93:8312-7. [PubMed]
- Jakob U, Muse W, Eser M, et al. Chaperone activity with a re-dox switch. Cell 1999;96:341-52. [PubMed]
- Ilbert M, Graf PC, Jakob U. Zinc center as redox switch--new function for an old motif. Antioxid Redox Signal 2006;8:835-46. [PubMed]
- Leonard TA, Rozycki B, Saidi LF, et al. Crystal structure and allosteric activation of protein kinase C betaII. Cell 2011;144:55-66. [PubMed]
- Korichneva I, Hoyos B, Chua R, et al. Zinc-release from protein kinase C as the common event during activation by lipid second messenger or reactive oxygen. J Biol Chem 2002;277:44327-31. [PubMed]
- Pelicci G, Lanfrancone L, Grignani F, et al. A novel transforming protein SHC) with an SH2 domain is implicated in mitogenic signal transduction. Cell 1992;70:93-104. [PubMed]
- Morita M, Matsuzaki H, Yamamoto T, et al. Epidermal growth factor receptor phosphorylates protein kinase C {delta} at Tyr332 to form a tri-meric complex with p66Shc in the H2O2-stimulated cells. J Biochem 2008;143:31-8. [PubMed]
- Giorgio M, Migliaccio E, Orsini F, et al. Electron Transfer be-tween Cytochrome c and p66Shc Generates Reactive Oxygen Species that Trigger Mitochondrial Apoptosis. Cell 2005;122:221-33. [PubMed]
- Lodish H, Berk A, Matsudaira P, et al, editors. Molecular Cell Biology. New York: W. H. Freeman and Company, 2004.
- Braiman L, Alt A, Kuroki T, et al. Protein kinase Cdelta mediates insulin-induced glucose transport in primary cultures of rat skeletal muscle. Mol Endocrinol 1999;13:2002-12. [PubMed]
- Uchida T, Iwashita N, Ohara-Imaizumi M, et al. Protein kinase Cdelta plays a non-redundant role in insulin secretion in pancreatic beta cells. J Biol Chem 2007;282:2707-16. [PubMed]
- Hennige AM, Ranta F, Heinzelmann I, et al. Overexpression of kinase-negative protein kinase Cdelta in pancreatic beta-cells protects mice from diet-induced glucose intolerance and beta-cell dysfunction. Diabetes 2010;59:119-27. [PubMed]
- Braiman L, Alt A, Kuroki T, et al. Insulin induces specific inter-action between insulin receptor and protein kinase C delta in primary cultured skeletal muscle. Mol Endocrinol 2001;15:565-74. [PubMed]
- Horovitz-Fried M, Jacob AI, Cooper DR, et al. Activation of the nuclear transcription factor SP-1 by insulin rapidly increases the expression of protein kinase C delta in skeletal muscle. Cell Signal 2007;19:556-62. [PubMed]
- Greene MW, Ruhoff MS, Roth RA, et al. PKCdelta-mediated IRS-1 Ser24 phosphorylation negatively regulates IRS-1 function. Biochem Biophys Res Commun 2006;349:976-86. [PubMed]
- Lam TK, Yoshii H, Haber CA, et al. Free fatty acid-induced hepatic insulin resistance: a potential role for protein kinase C-delta. Am J Physiol Endocrinol Metab 2002;283:E682-91. [PubMed]
- Majumder PK, Mishra NC, Sun X, et al. Targeting of protein kinase C delta to mitochondria in the oxidative stress response. Cell Growth Differ 2001;12:465-70. [PubMed]
- Nishikawa T, Edelstein D, Du XL, et al. Normalizing mito-chondrial superoxide production blocks three pathways of hyperglycaemic dam-age. Nature 2000;404:787-90. [PubMed]
- Talior I, Tennenbaum T, Kuroki T, et al. PKC-delta-dependent activation of oxidative stress in adipocytes of obese and insulin-resistant mice: role for NADPH oxidase. Am J Physiol Endocrinol Metab 2005;288:E405-11. [PubMed]
- Greene MW, Burrington CM, Lynch DT, et al. Lipid metabolism, oxidative stress and cell death are regulated by PKC delta in a dietary mod-el of nonalcoholic steatohepatitis. PLoS One 2014;9:e85848. [PubMed]
- Migliaccio E, Giorgio M, Mele S, et al. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature 1999;402:309-13. [PubMed]
- Migliaccio E, Giorgio M, Pelicci PG. Apoptosis and aging: role of p66Shc redox protein. Antioxid Redox Signal 2006;8:600-8. [PubMed]
- Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science 2005;307:384-7. [PubMed]
- Bezy O, Tran TT, Pihlajamaki J, et al. PKCdelta regulates he-patic insulin sensitivity and hepatosteatosis in mice and humans. J Clin Invest 2011;121:2504-17. [PubMed]
- Berniakovich I, Trinei M, Stendardo M, et al. p66Shc-generated oxidative signal promotes fat accumulation. J Biol Chem 2008;283:34283-93. [PubMed]
- Ranieri SC, Fusco S, Panieri E, et al. Mammalian life-span determinant p66shcA mediates obesity-induced insulin resistance. Proc Natl Acad Sci U S A 2010;107:13420-5. [PubMed]
- Decensi A, Zanardi S, Argusti A, et al. Fenretinide and risk re-duction of second breast cancer. Nat Clin Pract Oncol 2007;4:64-5. [PubMed]
- Johansson H, Gandini S, Guerrieri-Gonzaga A, et al. Effect of fenretinide and low-dose tamoxifen on insulin sensitivity in premenopausal women at high risk for breast cancer. Cancer Res 2008;68:9512-8. [PubMed]
- Preitner F, Mody N, Graham TE, et al. Long-term Fenretinide treatment prevents high-fat diet-induced obesity, insulin resistance, and hepatic steatosis. Am J Physiol Endocrinol Metab 2009;297:E1420-9. [PubMed]
- Yang Q, Graham TE, Mody N, et al. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature 2005;436:356-62. [PubMed]
- Quadro L, Blaner WS, Salchow DJ, et al. Impaired retinal function and vitamin A availability in mice lacking retinol-binding protein. EMBO J 1999;18:4633-44. [PubMed]
- Kawaguchi R, Yu J, Honda J, et al. A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science 2007;315:820-5. [PubMed]
- Berry DC, Jin H, Majumdar A, et al. Signaling by vitamin A and retinol-binding protein regulates gene expression to inhibit insulin responses. Proc Natl Acad Sci U S A 2011;108:4340-5. [PubMed]
- Berry DC, Noy N. Signaling by vitamin A and retinol-binding protein in regulation of insulin responses and lipid homeostasis. Biochim Biophys Acta 2012;1821:168-76.
- Berry DC, Jacobs H, Marwarha G, et al. The STRA6 receptor is essential for retinol-binding protein-induced insulin resistance but not for main-taining vitamin A homeostasis in tissues other than the eye. J Biol Chem 2013;288:24528-39. [PubMed]
- Moraes-Vieira PM, Yore MM, Dwyer PM, et al. RBP4 acti-vates antigen-presenting cells, leading to adipose tissue inflammation and sys-temic insulin resistance. Cell Metab 2014;19:512-26. [PubMed]
- Norseen J, Hosooka T, Hammarstedt A, et al. Retinol-binding protein 4 inhibits insulin signaling in adipocytes by inducing proinflammatory cytokines in macrophages through a c-Jun N-terminal kinase- and toll-like receptor 4-dependent and retinol-independent mechanism. Mol Cell Biol 2012;32:2010-9. [PubMed]
- Lotan R. Retinoids and apoptosis: implications for cancer chemo-prevention and therapy. J Natl Cancer Inst 1995;87:1655-7. [PubMed]
- O’Connell MJ, Chua R, Hoyos B, et al. Retro-retinoids in regu-lated cell growth and death. J Exp Med 1996;184:549-55. [PubMed]
- Chiu HJ, Fischman DA, Hammerling U. Vitamin A-depletion causes oxidative stress, mitochondrial dysfunction and PARP-1-dependent energy deprivation. FASEB J 2008;22:3878-87. [PubMed]
- Korichneva I, Waka J, Hammerling U. Regulation of the cardiac mitochondrial membrane potential by retinoids. J Pharmacol Exp Ther 2003;305:426-33. [PubMed]
- Hail N, Kim HJ, Lotan R. Mechanisms of fenretinide-induced apoptosis. Apoptosis 2006;11:1677-94. [PubMed]
- Noy N, Xu ZJ. Kinetic parameters of the interactions of reti-nol with lipid bilayers. Biochemistry 1990;29:3883-8. [PubMed]
- Berry DC, O'Byrne SM, Vreeland AC, et al. Cross talk be-tween signaling and vitamin A transport by the retinol-binding protein receptor STRA6. Mol Cell Biol 2012;32:3164-75. [PubMed]
- Noy N, Blaner WS. Interactions of retinol with binding proteins: studies with rat cellular retinol-binding protein and with rat retinol-binding protein. Biochemistry 1991;30:6380-6. [PubMed]


